如何开始
1、光标移动到下面的 import keras Cell中;
2、Shift+Enter或点击上面的运行按钮(类似播放)
出现Using Theano backend.那么Keras就已经成功安装了
import keras
#查看mnist数据集
%matplotlib inline
from keras.datasets import mnist
from matplotlib import pyplot as plt
# load data
(X_train, y_train), (X_test, y_test) = mnist.load_data("/notebook/datasets/mnist.pkl")
# create a grid of 3x3 images
for i in range(0, 9):
plt.subplot(330 + 1 + i)
plt.imshow(X_train[i], cmap=plt.get_cmap('gray'))
# show the plot
plt.show()
%matplotlib inline
from matplotlib import pyplot as plt
from keras.models import Sequential
from keras.layers import Dense,Dropout
from keras.models import model_from_json
import numpy
import os
# 为了多次执行再现结果,这只一个固定的随机数 fix random seed for reproducibility
seed = 7
numpy.random.seed(seed)
# 加载数据集 load pima indians dataset
dataset = numpy.loadtxt("pima-indians-diabetes.csv", delimiter=",")
# 分开数据集为输入和输出两部分 split into input (X) and output (Y) variables
X = dataset[:,0:8]
Y = dataset[:,8]
print (X.shape,Y.shape)
print (Y)
(768, 8) (768,)
[ 1. 0. 1. 0. 1. 0. 1. 0. 1. 1. 0. 1. 0. 1. 1. 1. 1. 1.
0. 1. 0. 0. 1. 1. 1. 1. 1. 0. 0. 0. 0. 1. 0. 0. 0. 0.
0. 1. 1. 1. 0. 0. 0. 1. 0. 1. 0. 0. 1. 0. 0. 0. 0. 1.
0. 0. 1. 0. 0. 0. 0. 1. 0. 0. 1. 0. 1. 0. 0. 0. 1. 0.
1. 0. 0. 0. 0. 0. 1. 0. 0. 0. 0. 0. 1. 0. 0. 0. 1. 0.
0. 0. 0. 1. 0. 0. 0. 0. 0. 1. 1. 0. 0. 0. 0. 0. 0. 0.
0. 1. 1. 1. 0. 0. 1. 1. 1. 0. 0. 0. 1. 0. 0. 0. 1. 1.
0. 0. 1. 1. 1. 1. 1. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 1.
0. 0. 0. 0. 0. 0. 0. 0. 1. 0. 1. 1. 0. 0. 0. 1. 0. 0.
0. 0. 1. 1. 0. 0. 0. 0. 1. 1. 0. 0. 0. 1. 0. 1. 0. 1.
0. 0. 0. 0. 0. 1. 1. 1. 1. 1. 0. 0. 1. 1. 0. 1. 0. 1.
1. 1. 0. 0. 0. 0. 0. 0. 1. 1. 0. 1. 0. 0. 0. 1. 1. 1.
1. 0. 1. 1. 1. 1. 0. 0. 0. 0. 0. 1. 0. 0. 1. 1. 0. 0.
0. 1. 1. 1. 1. 0. 0. 0. 1. 1. 0. 1. 0. 0. 0. 0. 0. 0.
0. 0. 1. 1. 0. 0. 0. 1. 0. 1. 0. 0. 1. 0. 1. 0. 0. 1.
1. 0. 0. 0. 0. 0. 1. 0. 0. 0. 1. 0. 0. 1. 1. 0. 0. 1.
0. 0. 0. 1. 1. 1. 0. 0. 1. 0. 1. 0. 1. 1. 0. 1. 0. 0.
1. 0. 1. 1. 0. 0. 1. 0. 1. 0. 0. 1. 0. 1. 0. 1. 1. 1.
0. 0. 1. 0. 1. 0. 0. 0. 1. 0. 0. 0. 0. 1. 1. 1. 0. 0.
0. 0. 0. 0. 0. 0. 0. 1. 0. 0. 0. 0. 0. 1. 1. 1. 0. 1.
1. 0. 0. 1. 0. 0. 1. 0. 0. 1. 1. 0. 0. 0. 0. 1. 0. 0.
1. 0. 0. 0. 0. 0. 0. 0. 1. 1. 1. 0. 0. 1. 0. 0. 1. 0.
0. 1. 0. 1. 1. 0. 1. 0. 1. 0. 1. 0. 1. 1. 0. 0. 0. 0.
1. 1. 0. 1. 0. 1. 0. 0. 0. 0. 1. 1. 0. 1. 0. 1. 0. 0.
0. 0. 0. 1. 0. 0. 0. 0. 1. 0. 0. 1. 1. 1. 0. 0. 1. 0.
0. 1. 0. 0. 0. 1. 0. 0. 1. 0. 0. 0. 0. 0. 0. 0. 0. 0.
1. 0. 0. 0. 0. 0. 0. 0. 1. 0. 0. 0. 1. 0. 0. 0. 1. 1.
0. 0. 0. 0. 0. 0. 0. 1. 0. 0. 0. 0. 1. 0. 0. 0. 1. 0.
0. 0. 1. 0. 0. 0. 1. 0. 0. 0. 0. 1. 1. 0. 0. 0. 0. 0.
0. 1. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 1. 0. 0. 0. 1.
1. 1. 1. 0. 0. 1. 1. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.
0. 0. 1. 1. 0. 0. 0. 0. 0. 0. 0. 1. 0. 0. 0. 0. 0. 0.
0. 1. 0. 1. 1. 0. 0. 0. 1. 0. 1. 0. 1. 0. 1. 0. 1. 0.
0. 1. 0. 0. 1. 0. 0. 0. 0. 1. 1. 0. 1. 0. 0. 0. 0. 1.
1. 0. 1. 0. 0. 0. 1. 1. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.
1. 0. 0. 0. 0. 1. 0. 0. 1. 0. 0. 0. 1. 0. 0. 0. 1. 1.
1. 0. 0. 0. 0. 0. 0. 1. 0. 0. 0. 1. 0. 1. 1. 1. 1. 0.
1. 1. 0. 0. 0. 0. 0. 0. 0. 1. 1. 0. 1. 0. 0. 1. 0. 1.
0. 0. 0. 0. 0. 1. 0. 1. 0. 1. 0. 1. 1. 0. 0. 0. 0. 1.
1. 0. 0. 0. 1. 0. 1. 1. 0. 0. 1. 0. 0. 1. 1. 0. 0. 1.
0. 0. 1. 0. 0. 0. 0. 0. 0. 0. 1. 1. 1. 0. 0. 0. 0. 0.
0. 1. 1. 0. 0. 1. 0. 0. 1. 0. 1. 1. 1. 0. 0. 1. 1. 1.
0. 1. 0. 1. 0. 1. 0. 0. 0. 0. 1. 0.]
%matplotlib inline
from matplotlib import pyplot as plt
from keras.models import Sequential
from keras.layers import Dense,Dropout
from keras.models import model_from_json
import numpy
import os
# 为了多次执行再现结果,这只一个固定的随机数 fix random seed for reproducibility
seed = 7
numpy.random.seed(seed)
# 加载数据集 load pima indians dataset
dataset = numpy.loadtxt("pima-indians-diabetes.csv", delimiter=",")
# 分开数据集为输入和输出两部分 split into input (X) and output (Y) variables
X = dataset[:,0:8]
Y = dataset[:,8]
# 创建模型 create model
model = Sequential()
model.add(Dense(12, input_dim=8, init='uniform', activation='relu'))
model.add(Dense(8, init='uniform', activation='relu'))
model.add(Dense(1, init='uniform', activation='sigmoid'))
# 编译模型 Compile model
model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])
# 训练模型 Fit the model
history= model.fit(X, Y,validation_split=0.25, nb_epoch=300, batch_size=10, verbose=0)
# 评估模型 evaluate the model
scores = model.evaluate(X, Y, verbose=0)
print("%s: %.2f%%" % (model.metrics_names[1], scores[1]*100))
# 保存模型 serialize model to JSON
model_json = model.to_json()
with open("./models/diabetes-model.json", "w") as json_file:
json_file.write(model_json)
# 保存权重 serialize weights to HDF5
model.save_weights("./models/diabetes-model.h5")
print("Saved model to disk")
#训练过程可视化
# list all data in history
print(history.history.keys())
# summarize history for accuracy
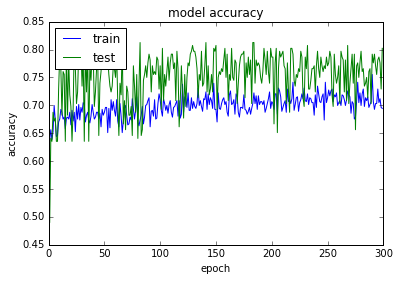
plt.plot(history.history['acc'])
plt.plot(history.history['val_acc'])
plt.title('model accuracy')
plt.ylabel('accuracy')
plt.xlabel('epoch')
plt.legend(['train', 'test'], loc='upper left')
plt.show()
# summarize history for loss
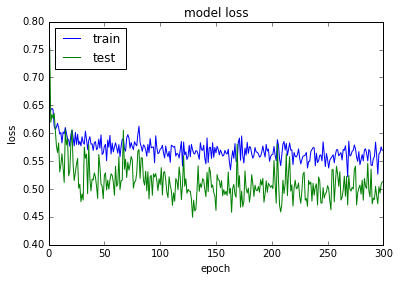
plt.plot(history.history['loss'])
plt.plot(history.history['val_loss'])
plt.title('model loss')
plt.ylabel('loss')
plt.xlabel('epoch')
plt.legend(['train', 'test'], loc='upper left')
plt.show()
acc: 80.08%
Saved model to disk
dict_keys(['val_acc', 'val_loss', 'loss', 'acc'])
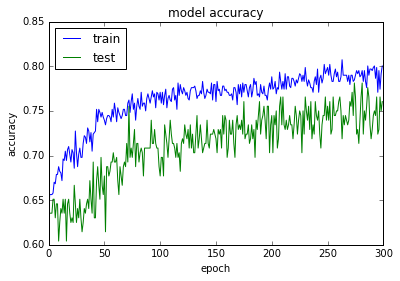
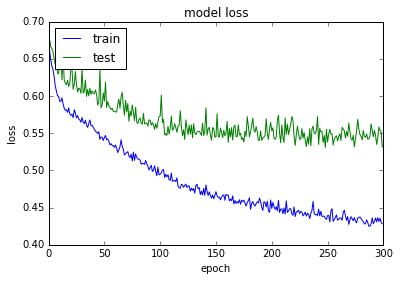
使用正则化和Dropout
%matplotlib inline
from matplotlib import pyplot as plt
from keras.models import Sequential
from keras.layers import Dense,Dropout,Activation
from keras.models import model_from_json
import numpy
import os
#正则化
# import BatchNormalization
from keras.layers.normalization import BatchNormalization
# 为了多次执行再现结果,这只一个固定的随机数 fix random seed for reproducibility
seed = 7
numpy.random.seed(seed)
# 加载数据集 load pima indians dataset
dataset = numpy.loadtxt("pima-indians-diabetes.csv", delimiter=",")
# 分开数据集为输入和输出两部分 split into input (X) and output (Y) variables
X = dataset[:,0:8]
Y = dataset[:,8]
# 创建模型 create model
model = Sequential()
model.add(Dense(12, input_dim=8, init='uniform'))
model.add(BatchNormalization())
model.add(Activation('relu'))
model.add(Dropout(0.5))
model.add(Dense(8, init='uniform'))
model.add(BatchNormalization())
model.add(Activation('relu'))
model.add(Dropout(0.5))
model.add(Dense(1, init='uniform'))
model.add(BatchNormalization())
model.add(Activation('sigmoid'))
# 编译模型 Compile model
model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])
# 训练模型 Fit the model
history= model.fit(X, Y,validation_split=0.25, nb_epoch=300, batch_size=10, verbose=0)
# 评估模型 evaluate the model
scores = model.evaluate(X, Y, verbose=0)
print("%s: %.2f%%" % (model.metrics_names[1], scores[1]*100))
# 保存模型 serialize model to JSON
model_json = model.to_json()
with open("./models/diabetes-model.json", "w") as json_file:
json_file.write(model_json)
# 保存权重 serialize weights to HDF5
model.save_weights("./models/diabetes-model.h5")
print("Saved model to disk")
#训练过程可视化
# list all data in history
print(history.history.keys())
# summarize history for accuracy
plt.plot(history.history['acc'])
plt.plot(history.history['val_acc'])
plt.title('model accuracy')
plt.ylabel('accuracy')
plt.xlabel('epoch')
plt.legend(['train', 'test'], loc='upper left')
plt.show()
# summarize history for loss
plt.plot(history.history['loss'])
plt.plot(history.history['val_loss'])
plt.title('model loss')
plt.ylabel('loss')
plt.xlabel('epoch')
plt.legend(['train', 'test'], loc='upper left')
plt.show()
acc: 76.69%
Saved model to disk
dict_keys(['val_acc', 'val_loss', 'loss', 'acc'])
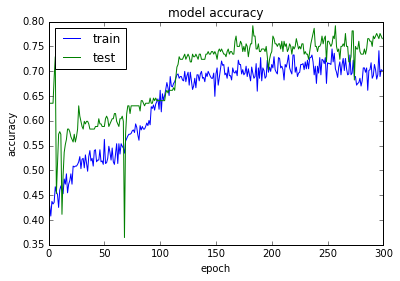
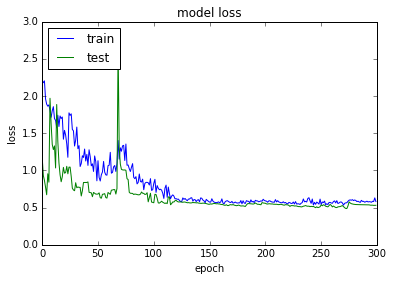
使用SGD优化器
%matplotlib inline
from matplotlib import pyplot as plt
from keras.models import Sequential
from keras.optimizers import SGD
from keras.layers import Dense,Dropout,Activation
from keras.models import model_from_json
import numpy
import os
#正则化
# import BatchNormalization
from keras.layers.normalization import BatchNormalization
# 为了多次执行再现结果,这只一个固定的随机数 fix random seed for reproducibility
seed = 7
numpy.random.seed(seed)
# 加载数据集 load pima indians dataset
dataset = numpy.loadtxt("pima-indians-diabetes.csv", delimiter=",")
# 分开数据集为输入和输出两部分 split into input (X) and output (Y) variables
X = dataset[:,0:8]
Y = dataset[:,8]
# 创建模型 create model
model = Sequential()
model.add(Dense(12, input_dim=8, init='uniform'))
model.add(BatchNormalization())
model.add(Activation('relu'))
model.add(Dropout(0.5))
model.add(Dense(8, init='uniform'))
model.add(BatchNormalization())
model.add(Activation('relu'))
model.add(Dropout(0.5))
model.add(Dense(1, init='uniform'))
model.add(BatchNormalization())
model.add(Activation('sigmoid'))
# 编译模型 Compile model
sgd = SGD(lr=0.1, decay=1e-6, momentum=0.9, nesterov=True)
model.compile(loss='binary_crossentropy', optimizer=sgd, metrics=['accuracy'])
# 训练模型 Fit the model
history= model.fit(X, Y,validation_split=0.25, nb_epoch=300, batch_size=10, verbose=0)
# 评估模型 evaluate the model
scores = model.evaluate(X, Y, verbose=0)
print("%s: %.2f%%" % (model.metrics_names[1], scores[1]*100))
# 保存模型 serialize model to JSON
model_json = model.to_json()
with open("./models/diabetes-model.json", "w") as json_file:
json_file.write(model_json)
# 保存权重 serialize weights to HDF5
model.save_weights("./models/diabetes-model.h5")
print("Saved model to disk")
#训练过程可视化
# list all data in history
print(history.history.keys())
# summarize history for accuracy
plt.plot(history.history['acc'])
plt.plot(history.history['val_acc'])
plt.title('model accuracy')
plt.ylabel('accuracy')
plt.xlabel('epoch')
plt.legend(['train', 'test'], loc='upper left')
plt.show()
# summarize history for loss
plt.plot(history.history['loss'])
plt.plot(history.history['val_loss'])
plt.title('model loss')
plt.ylabel('loss')
plt.xlabel('epoch')
plt.legend(['train', 'test'], loc='upper left')
plt.show()
acc: 76.69%
Saved model to disk
dict_keys(['val_acc', 'val_loss', 'loss', 'acc'])
%matplotlib inline
from matplotlib import pyplot as plt
from keras.callbacks import EarlyStopping,ModelCheckpoint
from keras.models import Sequential
from keras.layers import Dense,Dropout
from keras.models import model_from_json
import numpy as np
import os
# 为了多次执行再现结果,这只一个固定的随机数 fix random seed for reproducibility
seed = 7
np.random.seed(seed)
# 加载数据集 load pima indians dataset
dataset = numpy.loadtxt("pima-indians-diabetes.csv", delimiter=",")
# 分开数据集为输入和输出两部分 split into input (X) and output (Y) variables
X = dataset[:,0:8]
Y = dataset[:,8]
# 创建模型 create model
model = Sequential()
model.add(Dense(12, input_dim=8, init='uniform', activation='relu'))
model.add(Dense(8, init='uniform', activation='relu'))
model.add(Dense(1, init='uniform', activation='sigmoid'))
# 编译模型 Compile model
model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])
#--早停 和检查点-callback---
filepath="./temp/diabetes-weights-improvement-{epoch:02d}-{val_acc:.2f}.hdf5"
checkpoint = ModelCheckpoint(filepath, monitor='val_acc', verbose=1, save_best_only=True, mode='max')
checkpoint2=ModelCheckpoint("./temp/diabetes-weights.{epoch:02d}-{val_loss:.2f}.h5", monitor='val_loss', verbose=0, save_best_only=False, mode='auto')
#当监测值不再改善时,该回调函数将中止训练
# patience:当early stop被激活(如发现loss相比上一个epoch训练没有下降),
# 则经过patience个epoch后停止训练。
estop = EarlyStopping(monitor='val_loss', patience=5, verbose=0, mode='auto')
#--------------------------------------
# 训练模型 Fit the model
history= model.fit(X, Y,validation_split=0.5, nb_epoch=150, batch_size=10, verbose=0,callbacks=[checkpoint,checkpoint2,estop])
# 评估模型 evaluate the model
scores = model.evaluate(X, Y, verbose=0)
print("%s: %.2f%%" % (model.metrics_names[1], scores[1]*100))
# 保存模型 serialize model to JSON
model_json = model.to_json()
with open("./models/diabetes-model.json", "w") as json_file:
json_file.write(model_json)
# 保存权重 serialize weights to HDF5
model.save_weights("./models/diabetes-model.h5")
print("Saved model to disk")
#训练过程可视化
# list all data in history
print(history.history.keys())
# summarize history for accuracy
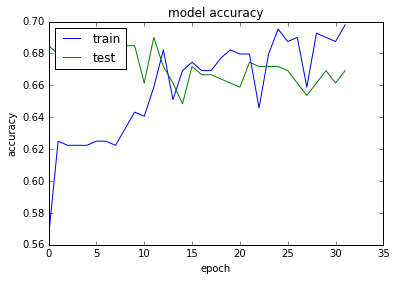
plt.plot(history.history['acc'])
plt.plot(history.history['val_acc'])
plt.title('model accuracy')
plt.ylabel('accuracy')
plt.xlabel('epoch')
plt.legend(['train', 'test'], loc='upper left')
plt.show()
# summarize history for loss
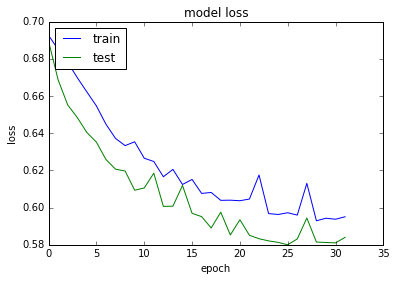
plt.plot(history.history['loss'])
plt.plot(history.history['val_loss'])
plt.title('model loss')
plt.ylabel('loss')
plt.xlabel('epoch')
plt.legend(['train', 'test'], loc='upper left')
plt.show()
Epoch 00000: val_acc improved from -inf to 0.68490, saving model to ./temp/diabetes-weights-improvement-00-0.68.hdf5
Epoch 00001: val_acc did not improve
Epoch 00002: val_acc did not improve
Epoch 00003: val_acc did not improve
Epoch 00004: val_acc did not improve
Epoch 00005: val_acc did not improve
Epoch 00006: val_acc improved from 0.68490 to 0.69010, saving model to ./temp/diabetes-weights-improvement-06-0.69.hdf5
Epoch 00007: val_acc did not improve
Epoch 00008: val_acc did not improve
Epoch 00009: val_acc did not improve
Epoch 00010: val_acc did not improve
Epoch 00011: val_acc improved from 0.69010 to 0.69010, saving model to ./temp/diabetes-weights-improvement-11-0.69.hdf5
Epoch 00012: val_acc did not improve
Epoch 00013: val_acc did not improve
Epoch 00014: val_acc did not improve
Epoch 00015: val_acc did not improve
Epoch 00016: val_acc did not improve
Epoch 00017: val_acc did not improve
Epoch 00018: val_acc did not improve
Epoch 00019: val_acc did not improve
Epoch 00020: val_acc did not improve
Epoch 00021: val_acc did not improve
Epoch 00022: val_acc did not improve
Epoch 00023: val_acc did not improve
Epoch 00024: val_acc did not improve
Epoch 00025: val_acc did not improve
Epoch 00026: val_acc did not improve
Epoch 00027: val_acc did not improve
Epoch 00028: val_acc did not improve
Epoch 00029: val_acc did not improve
Epoch 00030: val_acc did not improve
Epoch 00031: val_acc did not improve
acc: 69.01%
Saved model to disk
dict_keys(['val_acc', 'val_loss', 'loss', 'acc'])
loss, accuracy = model.evaluate(X_test,Y_test, verbose=0)
predicted_classes = model.predict_classes(X_test)
correct_classified_indices = np.nonzero(predicted_classes == y_test)[0]
incorrect_classified_indices = np.nonzero(predicted_classes != y_test)[0]
correct_classified_indices
array([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 14, 15, 16])
incorrect_classified_indices
array([ 0, 13])







